Creates a graph using the climate and elevation data which has
been extracted for a given location. It accepts the data formatted
from the ce_extract function.
plot_c(
data,
geo_id,
interval_prec = 20,
interval_temp = 5,
stretch_temp = 4,
p_cex = 1,
nchr_main = 32,
l_main = 0.1,
l_prec = 0,
l_month = -0.6,
lwd_temp = 1.5,
lwd_prec = 9,
l_units = 0.1,
l_tcols = c(14.5, 16.5, 18.5, 19.5, 22.5)
)Arguments
- data
List. Containing climate, elevation and latitude data sets. Structured by
ce_extract().- geo_id
Character. Corresponding to a specific feature contained in the
"location"argument (specifically in the"location_g"column).- interval_prec
Numeric. Interval of the precipitation axis.
- interval_temp
Numeric. Interval of the temperature axis.
- stretch_temp
Numeric. Ratio for stretching temperature relative to precipitation.
- p_cex
Numeric. Text size for entire plot.
- nchr_main
Numeric. Controls the number of characters of the main title before wrapping to a new line.
- l_main
Numeric. Line position of the main title.
- l_prec
Numeric. Line position of the y axis (precipitation).
- l_month
Numeric. Line position of the x axis (months).
- lwd_temp
Numeric. Line width of temperature.
- lwd_prec
Numeric. Line width of precipitation.
- l_units
Numeric. Line position of precipitation and temperature units.
- l_tcols
List. Line position of the table columns. Must be length 5 corresponding to the position (left to right) for each column.
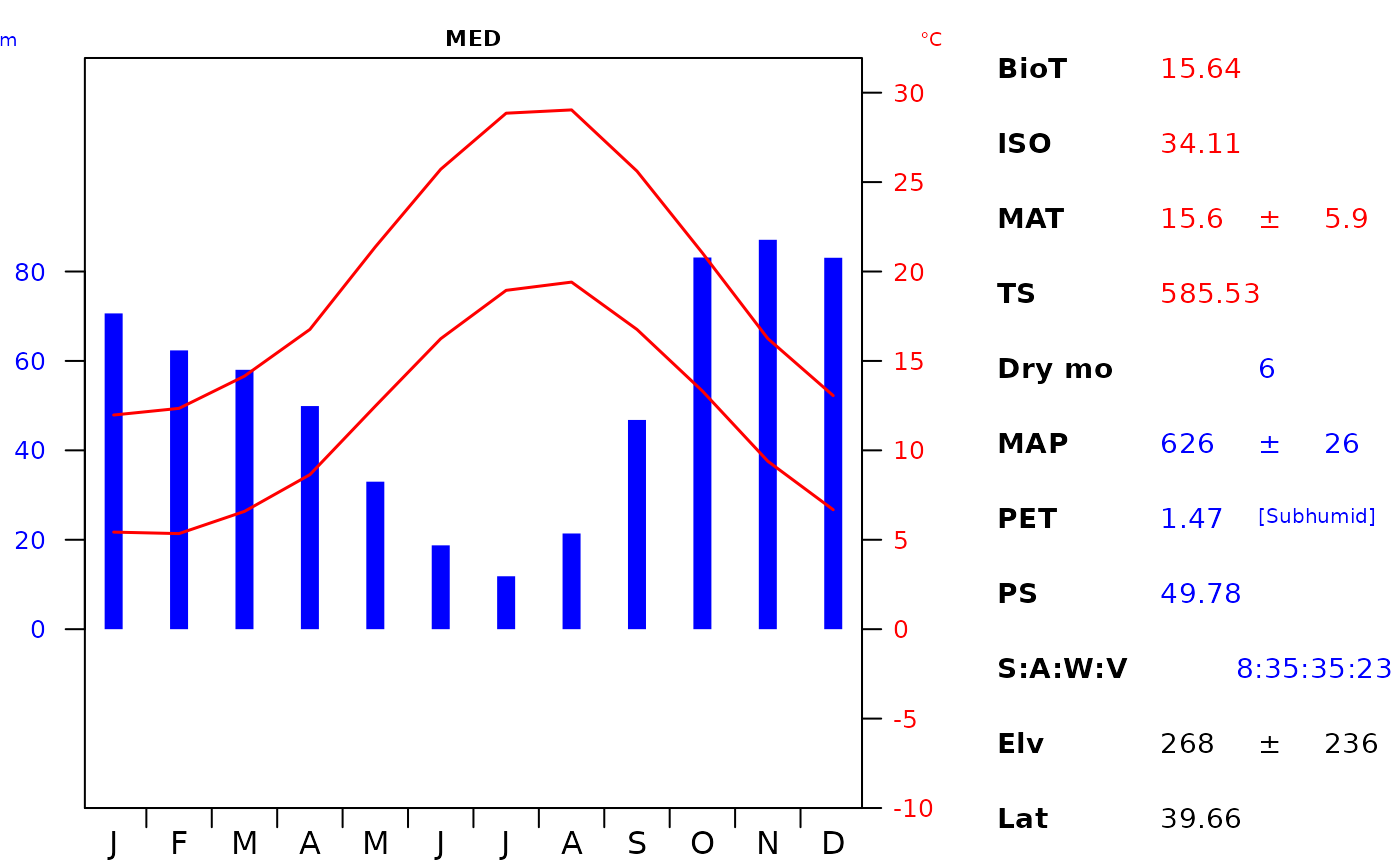
Value
Returns a base R family of plot. This function uses the dismo package to calculate isothermality (ISO), temperature seasonality (TS) and precipitation seasonality (PS).
References
Pizarro, M, Hernangómez, D. & Fernández-Avilés G. (2023). climaemet: Climate AEMET Tools. Comprehensive R Archive Network. doi:10.5281/zenodo.5205573
Walter, H.B., & Lieth, H. (1960). Klimadiagramm-Weltatlas. VEB Gustav Fischer Verlag, Jena.
See also
Download climate data: ce_download()
Examples
# Step 1. Import the Italian Biome polygon data
# Step 2. Run the download function
# Step 3. Run the extract function
#* See ce_download & ce_extract documentation
# Steps 1, 2 & 3 can be skipped by loading the extracted data (it_data)
data("it_data", package = "climenv")
# Step 4. Visualise the climatic envelope using our custom diagram
# first lets store the default graphics parameters
# we need to make some changes to ensure that the table fits in the plotting
# region.
# Set up plotting parameters
oldpar <- par(mar = c(1.5, 2.2, 1.5, 14) + 0.01)
plot_c(
it_data, geo_id = "MED",
l_tcols = c(14.5, 17, 18.5, 19.5, 21)
)
 # Restore user options
par(oldpar)
# This output works if you export to a three column width sized image.
# Restore user options
par(oldpar)
# This output works if you export to a three column width sized image.